from fastai.vision.all import *Lesson Video:
What is Semantic Segmentation?
Given a particular image, we can assume that each pixel inside that image represents a class.
This can come in the form (most commonly) of:
- Binary: either it is or is not (such as background v.s. an object)
- Multiclass: one of
nnumber of classes (such as parts of the body)
In the Practical Deep Learning for Coders course it was the latter shown through the CAMVID problem. In this course we will focus on the binary segmentation aspect as sometimes it takes a bit of preprocessing to get it going well.
The other option would be when individual pixels represent multiple classes, though I haven’t seen too many of these cases personally.
The Dataset
The dataset for today will be portraits of people. Let’s download it first.
Make sure to run pip install gdown first, as the dataset is hosted on Google Drive!
url = "https://drive.google.com/uc?id=18xM3jU2dSp1DiDqEM6PVXattNMZvsX4z"!gdown {url}Downloading...
From: https://drive.google.com/uc?id=18xM3jU2dSp1DiDqEM6PVXattNMZvsX4z
To: /content/Portrait.zip
100% 107M/107M [00:04<00:00, 23.3MB/s] When working in Jupyter Notebooks you can utilize variables named earlier when calling bash commands by surrounding them with {}
The data has now been stored in a Portrait.zip file. Let’s use the zipfile library to unzip it to a data folder:
from zipfile import ZipFile
with ZipFile("Portrait.zip", "r") as zip_ref:
zip_ref.extractall("data")What’s inside?
path = Path("data")for walk in path.ls():
print(repr(walk), walk.is_file())Path('../data/images.csv') True
Path('../data/images_compressed') False
Path('../data/images_original') False
Path('../data/images_data_crop') False
Path('../data/train.txt') True
Path('../data/GT_png') False
Path('../data/EG1800_val.txt') True
Path('../data/val.txt') True
Path('../data/EG1800_train.txt') TrueYou can see that we have some files relating to image labels, filenames that relate to whether an image is in the train or validation set, and two other text files we won’t worry about.
Checking the Labels
Why check the labels next?
Let’s grab one and take a look:
(path/"GT_png").ls()[0]Path('../data/GT_png/02218_mask.png')mask = Image.open((path/"GT_png").ls()[0])mask
In semantic segmentation, the “labels” are a 1:1 mask of the original picture with each pixel representing a label and are single channel:
mask = np.asarray(mask); maskarray([[ 0, 0, 0, ..., 0, 0, 0],
[ 0, 0, 0, ..., 0, 0, 0],
[ 0, 0, 0, ..., 0, 0, 0],
...,
[255, 255, 255, ..., 0, 0, 0],
[255, 255, 255, ..., 0, 0, 0],
[255, 255, 255, ..., 0, 0, 0]], dtype=uint8)Very quickly we see the issue!
Because of how the loss gets calculated (and how fastai does things in general), the values of the pixel mask must be from 0 -> n, with n being the number of classes possible. If we take things as they are here during training you’ll hit an error that says “CUDA Segmentation Fault, Index Out of Bounds” (or something similar).
This is because our labels should be from 0 -> 1, to align with the fact predicted probabilities from our model are 0 -> 1. Instead they are 0 and 255, leading to this issue.
So how do we fix the issue? In numpy we can just override the numbers for a particular value in the array and set it. To generalize this however a dictionary of the original value to the new one should also be made:
def get_codes(fnames) -> Dict[int,int]:
"Returns a dictionary of `original_code:new_code` for pixel values in segmentation masks"
unique_codes = set()
for fname in fnames:
mask = Image.open(fname)
mask = np.asarray(mask)
for color in np.unique(mask):
unique_codes.add(color)
return {
i : color
for i, color in
enumerate(unique_codes)
}def get_codes(fnames) -> Dict[int,int]:
"Returns a dictionary of `original_code:new_code` for pixel values in segmentation masks"
unique_codes = set()
for fname in fnames:
mask = Image.open(fname)
mask = np.asarray(mask)
for color in np.unique(mask):
unique_codes.add(color)
return {
i : color
for i, color in
enumerate(unique_codes)
}def get_codes(fnames) -> Dict[int,int]: The function will take in a list of filenames and return a dictionary of int:int
unique_codes = set()The unique pixel values found in each mask will be stored in a set called unique_codes. This is because a set let’s us add items to the object but it will only be included if it does not already exist inside it.
mask = Image.open(fname)
mask = np.asarray(mask)We need to open each mask and convert it to a numpy array
for color in np.unique(mask):
unique_codes.add(color)np.unique will find all the different pixel values present in our mask and return them as an array. We need to then add each of them to our set to see what ones exist.
return {
i : color
for i, color in
enumerate(unique_codes)
}Finally, we can construct a dictionary from 0 to n that says what the pixel to color translation will be.
unique_codes = get_codes((path/"GT_png").ls()[:20])
unique_codes{0: 0, 1: 255}Changing the values inside our masks afterwards looks something like so:
mask = mask.copy()
np.place(mask, mask==255, 1)
np.unique(mask)array([0, 1], dtype=uint8)mask = mask.copy()
np.place(mask, mask==255, 1)
np.unique(mask)array([0, 1], dtype=uint8)mask = mask.copy()numpy will raise a WRITEBACKIFCOPY issue if we try to modify the original array in-place, so we make another one instead.
np.place(mask, mask==255, 1)np.place takes in an array, a mask, and values to replace them with. In this case the “mask” here means what values need changing.
We’ll need to perform this when we do our get_y
Building the Dataloaders
First step is to build a DataBlock (the Datasets will be shown afterwards). Our input is an Image and our output is a Mask, so thus:
codes = ["Background", "Face"]
blocks = (ImageBlock, MaskBlock(codes=codes))Codes are how fastai knows the human label for each image, so that when decoding the final output it isn’t an array of [0,1,0,...] but instead [Background, Face, Background,...]
Next we need to create a get_y function. In this case it should take in a filename and our dictionary, open the filename, and return the mask:
unique_codes{0: 0, 1: 255}def get_y(filename:Path, unique_codes:dict):
"Grabs a mask from `filename` and adjusts the pixel values based on `unique_codes`"
filename = path/"GT_png"/f'{filename.stem}_mask.png'
mask = np.asarray(Image.open(filename)).copy()
for new_value, old_value in unique_codes.items():
np.place(mask, mask==old_value, new_value)
return PILMask.create(mask)def get_y(filename:Path, unique_codes:dict):
"Grabs a mask from `filename` and adjusts the pixel values based on `unique_codes`"
filename = path/"GT_png"/f'{filename.stem}_mask.png'
mask = np.asarray(Image.open(filename)).copy()
for new_value, old_value in unique_codes.items():
np.place(mask, mask==old_value, new_value)
return PILMask.create(mask) filename = path/"GT_png"/f'{filename.stem}_mask.png'We need to grab the label relative to our x filename. It’s located under GT_png/{filename}_mask.png. stem is used to extract everything prior to the extension.
mask = np.asarray(Image.open(filename)).copy()Next similar to earlier we open the mask and convert it to a numpy array.
for new_value, old_value in unique_codes.items():
np.place(mask, mask==old_value, new_value)Then we replace all of the old values with the new ones
return PILMask.create(mask)Finally returning a fully created PILMask ready for fastai.
Now let’s test out if it works by calling the PILMask.show() function and passing in a color map to use
new_mask = get_y((path/"images_data_crop").ls()[0], unique_codes)
new_mask.show(cmap="Blues");
cmap relates to matplotlib and how they render colors for numerical values. Find out more here
DataBlock
We have a mask! Now everything is in place to build our DataBlock:
block = DataBlock(
blocks=blocks,
splitter=RandomSplitter(),
get_y=partial(get_y, unique_codes=unique_codes),
item_tfms=Resize(224),
batch_tfms=[*aug_transforms(), Normalize.from_stats(*imagenet_stats)]
)dls = block.dataloaders(
get_image_files(path/'images_data_crop'),
bs=8
)The architecture we will use for training (and UNet’s in general) are resource hungry, and we’re expecting a very large output so the batch sizes will need to be quite small.
dls.show_batch(cmap="Blues", vmin=0, vmax=1)
With Datasets
splitter = RandomSplitter()
dsets = Datasets(
get_image_files(path/'images_data_crop'),
tfms=[
[PILImage.create],
[partial(get_y, unique_codes=unique_codes)]
],
splits = splitter(get_image_files(path/'images_data_crop'))
)dls = dsets.dataloaders(
after_item = [
Resize(224),
ToTensor(),
AddMaskCodes(codes=codes)
],
after_batch = [
*aug_transforms(),
IntToFloatTensor(),
Normalize.from_stats(*imagenet_stats)
],
bs=8
)AddMaskCodes will inject that list of classes into the metadata of the ending TensorMask that is created. fastai will then use this data later when decoding all their values.
dls.show_batch(cmap="Blues", vmin=0, vmax=1)
With that we’re ready for training!
The U-Net
Insert scary noises here
When it comes to image segmentation, the model of choice with fastai is the Dynamic Unet.
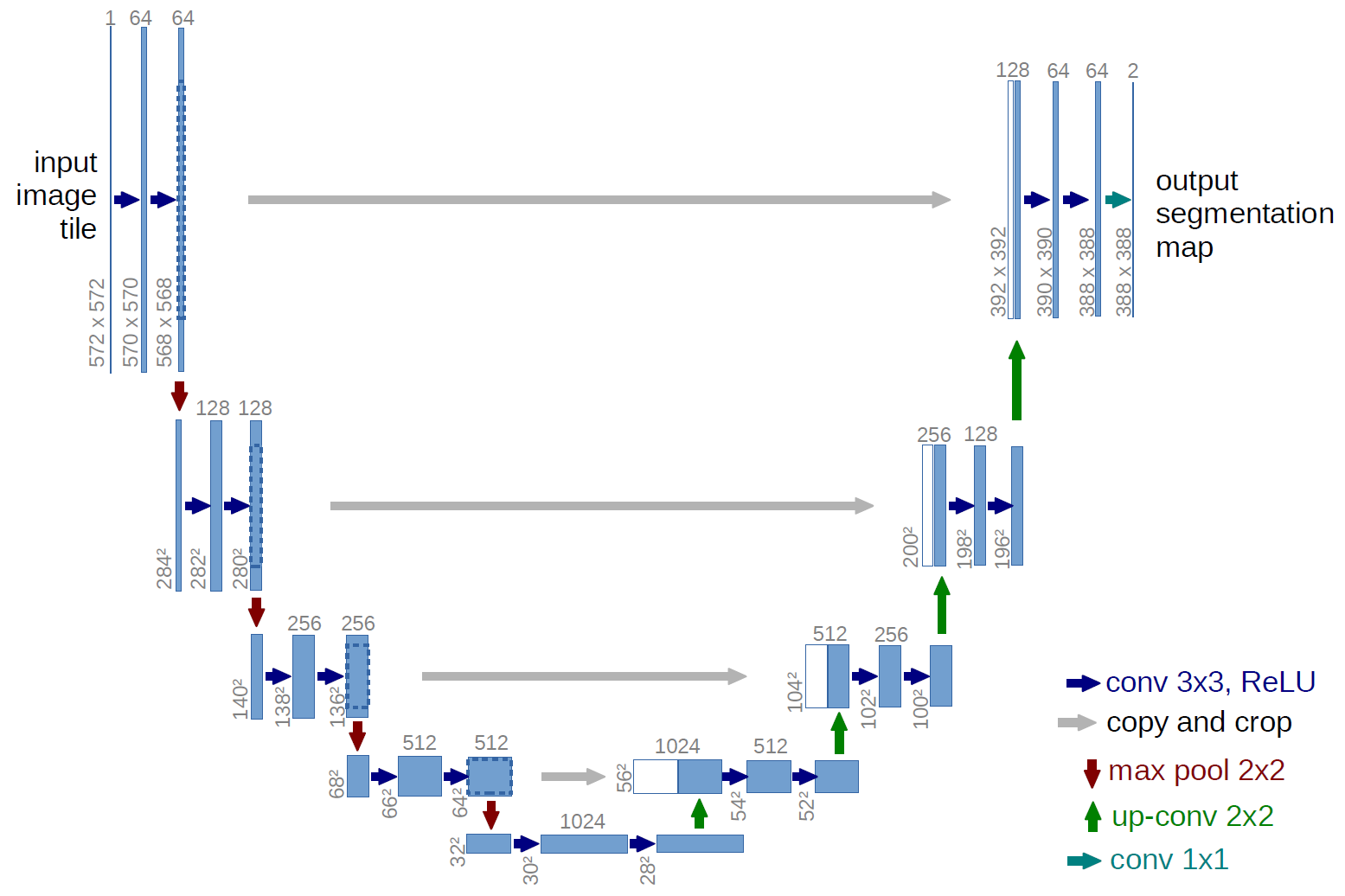
You may have seen places where the Unet is already being used, such as in Stable Diffusion. (We’re not going to talk about that here, feel free to go watch Jeremy’s course to learn more about that once it’s out!)
I highly recommend this video explaining how the UNet works!
Here’s the basic premise though. We take in these pixeled images of some size and shrink it down in half each time (encoding). Eventually it becomes a representation of some shape, in this case a matrix of 1024x30x30 (CxWxH). Afterwards we blow the image back up to it’s original size again (decoding) and create a segmentation map of size NxWxH where n is the number of classes, and W,H are the original width and height of the image when it was passed in.
Each channel in this final segmentation map represents all the values for a particular class as a result.
In fastai there is a special Learner function to create this model, unet_learner. There’s a variety of configurations we can pass to the unet, which you can read more about here however in my opinion the default configurations will get us by just fine.
The only two options we will be enabling are self attention layers (think EfficientNet or ViT) and changing the activation function to Mish, a popular alternative to ReLU:
learn = unet_learner(
dls,
resnet34,
metrics=partial(accuracy, axis=1),
self_attention=True,
act_cls=Mish,
loss_func = CrossEntropyLossFlat(axis=1)
)learn.summary()DynamicUnet (Input shape: 8 x 3 x 224 x 224)
============================================================================
Layer (type) Output Shape Param # Trainable
============================================================================
8 x 64 x 112 x 112
Conv2d 9408 False
BatchNorm2d 128 True
ReLU
____________________________________________________________________________
8 x 64 x 56 x 56
MaxPool2d
Conv2d 36864 False
BatchNorm2d 128 True
ReLU
Conv2d 36864 False
BatchNorm2d 128 True
Conv2d 36864 False
BatchNorm2d 128 True
ReLU
Conv2d 36864 False
BatchNorm2d 128 True
Conv2d 36864 False
BatchNorm2d 128 True
ReLU
Conv2d 36864 False
BatchNorm2d 128 True
____________________________________________________________________________
8 x 128 x 28 x 28
Conv2d 73728 False
BatchNorm2d 256 True
ReLU
Conv2d 147456 False
BatchNorm2d 256 True
Conv2d 8192 False
BatchNorm2d 256 True
Conv2d 147456 False
BatchNorm2d 256 True
ReLU
Conv2d 147456 False
BatchNorm2d 256 True
Conv2d 147456 False
BatchNorm2d 256 True
ReLU
Conv2d 147456 False
BatchNorm2d 256 True
Conv2d 147456 False
BatchNorm2d 256 True
ReLU
Conv2d 147456 False
BatchNorm2d 256 True
____________________________________________________________________________
8 x 256 x 14 x 14
Conv2d 294912 False
BatchNorm2d 512 True
ReLU
Conv2d 589824 False
BatchNorm2d 512 True
Conv2d 32768 False
BatchNorm2d 512 True
Conv2d 589824 False
BatchNorm2d 512 True
ReLU
Conv2d 589824 False
BatchNorm2d 512 True
Conv2d 589824 False
BatchNorm2d 512 True
ReLU
Conv2d 589824 False
BatchNorm2d 512 True
Conv2d 589824 False
BatchNorm2d 512 True
ReLU
Conv2d 589824 False
BatchNorm2d 512 True
Conv2d 589824 False
BatchNorm2d 512 True
ReLU
Conv2d 589824 False
BatchNorm2d 512 True
Conv2d 589824 False
BatchNorm2d 512 True
ReLU
Conv2d 589824 False
BatchNorm2d 512 True
____________________________________________________________________________
8 x 512 x 7 x 7
Conv2d 1179648 False
BatchNorm2d 1024 True
ReLU
Conv2d 2359296 False
BatchNorm2d 1024 True
Conv2d 131072 False
BatchNorm2d 1024 True
Conv2d 2359296 False
BatchNorm2d 1024 True
ReLU
Conv2d 2359296 False
BatchNorm2d 1024 True
Conv2d 2359296 False
BatchNorm2d 1024 True
ReLU
Conv2d 2359296 False
BatchNorm2d 1024 True
BatchNorm2d 1024 True
ReLU
____________________________________________________________________________
8 x 1024 x 7 x 7
Conv2d 4719616 True
Mish
____________________________________________________________________________
8 x 512 x 7 x 7
Conv2d 4719104 True
Mish
____________________________________________________________________________
8 x 1024 x 7 x 7
Conv2d 525312 True
Mish
____________________________________________________________________________
8 x 256 x 14 x 14
PixelShuffle
BatchNorm2d 512 True
Conv2d 2359808 True
Mish
Conv2d 2359808 True
Mish
Mish
____________________________________________________________________________
8 x 1024 x 14 x 14
Conv2d 525312 True
Mish
____________________________________________________________________________
8 x 256 x 28 x 28
PixelShuffle
BatchNorm2d 256 True
Conv2d 1327488 True
Mish
Conv2d 1327488 True
Mish
____________________________________________________________________________
8 x 48 x 784
Conv1d 18432 True
Conv1d 18432 True
Conv1d 147456 True
Mish
____________________________________________________________________________
8 x 768 x 28 x 28
Conv2d 295680 True
Mish
____________________________________________________________________________
8 x 192 x 56 x 56
PixelShuffle
BatchNorm2d 128 True
Conv2d 590080 True
Mish
Conv2d 590080 True
Mish
Mish
____________________________________________________________________________
8 x 512 x 56 x 56
Conv2d 131584 True
Mish
____________________________________________________________________________
8 x 128 x 112 x 112
PixelShuffle
BatchNorm2d 128 True
____________________________________________________________________________
8 x 96 x 112 x 112
Conv2d 165984 True
Mish
Conv2d 83040 True
Mish
Mish
____________________________________________________________________________
8 x 384 x 112 x 112
Conv2d 37248 True
Mish
____________________________________________________________________________
8 x 96 x 224 x 224
PixelShuffle
ResizeToOrig
____________________________________________________________________________
8 x 99 x 224 x 224
MergeLayer
Conv2d 88308 True
Mish
Conv2d 88308 True
Sequential
Mish
____________________________________________________________________________
8 x 2 x 224 x 224
Conv2d 200 True
ToTensorBase
____________________________________________________________________________
Total params: 41,405,488
Total trainable params: 20,137,840
Total non-trainable params: 21,267,648
Optimizer used: <function Adam>
Loss function: FlattenedLoss of CrossEntropyLoss()
Model frozen up to parameter group #2
Callbacks:
- TrainEvalCallback
- CastToTensor
- Recorder
- ProgressCallbackYou can see this change in shape visually in the architecture summary.
Otherwise training looks just as straightforward as before. Train our frozen backbone for a while:
learn.fit_one_cycle(10, 1e-3)| epoch | train_loss | valid_loss | accuracy | time |
|---|---|---|---|---|
| 0 | 0.152699 | 0.114538 | 0.960078 | 00:18 |
| 1 | 0.136885 | 0.101154 | 0.966822 | 00:16 |
| 2 | 0.097801 | 0.080650 | 0.973640 | 00:17 |
| 3 | 0.098249 | 0.092544 | 0.972757 | 00:17 |
| 4 | 0.083918 | 0.069728 | 0.975000 | 00:16 |
| 5 | 0.068980 | 0.065475 | 0.976745 | 00:17 |
| 6 | 0.072801 | 0.059130 | 0.979125 | 00:16 |
| 7 | 0.058030 | 0.057599 | 0.978940 | 00:16 |
| 8 | 0.057075 | 0.055490 | 0.979533 | 00:17 |
| 9 | 0.054590 | 0.055697 | 0.979628 | 00:17 |
Save the model in case something crashes:
learn.save("stage_1")
#learn.load("stage_1")Take a look at some results:
learn.show_results(max_n=4, figsize=(12,6))
Then unfreeze and train some more!
learn.unfreeze()
learn.fit_one_cycle(4, slice(1e-3/400, 1e-3/4))| epoch | train_loss | valid_loss | accuracy | time |
|---|---|---|---|---|
| 0 | 0.054487 | 0.057796 | 0.979340 | 00:17 |
| 1 | 0.061175 | 0.060088 | 0.978260 | 00:17 |
| 2 | 0.054855 | 0.055567 | 0.980046 | 00:17 |
| 3 | 0.046733 | 0.054867 | 0.980414 | 00:17 |
And look at what we have:
learn.show_results(max_n=4, figsize=(12,6))
Performing inference
Lastly we need to do some inference! We’ll start with using the test_dl and get_preds then similar to lesson 2 we’ll try just using fastai transforms and do it ourselves.
With test_dl
First we need to create a test dataloader using some filenames:
dl = learn.dls.test_dl(
(path/'images_data_crop').ls()[:5]
)
dl.show_batch()
dl = learn.dls.test_dl(
(path/'images_data_crop').ls()[:5]
)
dl.show_batch()
test_dl(...)The test_dl function will create a new DataLoader object based on the validation transforms in dls based on the data passed into it.
Then we can gather some predictions:
preds = learn.get_preds(dl=dl)If we check their shape:
preds[0].shapetorch.Size([5, 2, 224, 224])We’ll see that it’s NxCxWxH, as I mentioned earlier. But how do we fit this into a single-channel mask?
By argmax’ing the values on the first dimension (0):
preds[0][0].shapetorch.Size([2, 224, 224])pred = preds[0][0].argmax(dim=0)pred.shapetorch.Size([224, 224])We can then plot it to see our finished mask:
plt.imshow(pred);
Finally we need a way to save these results:
pred = pred.numpy()
rescaled = (255.0 / pred.max() * (pred - pred.min())).astype(np.uint8)
im = Image.fromarray(rescaled)
im.save("mask.png")pred = pred.numpy()
rescaled = (255.0 / pred.max() * (pred - pred.min())).astype(np.uint8)
im = Image.fromarray(rescaled)
im.save("mask.png")pred = pred.numpy()First to get away from tensors let’s convert the mask to a numpy array
rescaled = (255.0 / pred.max() * (pred - pred.min())).astype(np.uint8)Then we need to unscale the pixel values (as in de-normalize them).
im = Image.fromarray(rescaled)
im.save("mask.png")Finally we can create a Image from the array and save it!
im
Without test_dl
Now let’s perform the same with using our transforms:
fnames = (path/'images_data_crop').ls()[:5]
item_tfms = Pipeline([
PILImage.create,
RandomResizedCrop(224),
ToTensor()
], split_idx=1)
batch_tfms = Pipeline([
IntToFloatTensor(),
Normalize.from_stats(*imagenet_stats)
])
batch = []
for fname in fnames:
batch.append(item_tfms(fname))
batch = torch.stack(batch, dim=0)
batch = batch_tfms(batch.cuda())
model = learn.model
model.eval()
with torch.no_grad():
preds = model(batch)
for i,pred in enumerate(preds):
pred = pred.argmax(0)
pred = pred.cpu().numpy()
rescaled = (255.0 / pred.max() * (pred - pred.min())).astype(np.uint8)
im = Image.fromarray(rescaled)
im.save(f'pred_{i}.png')fnames = (path/'images_data_crop').ls()[:5]
item_tfms = Pipeline([
PILImage.create,
RandomResizedCrop(224),
ToTensor()
], split_idx=1)
batch_tfms = Pipeline([
IntToFloatTensor(),
Normalize.from_stats(*imagenet_stats)
])
batch = []
for fname in fnames:
batch.append(item_tfms(fname))
batch = torch.stack(batch, dim=0)
batch = batch_tfms(batch.cuda())
model = learn.model
model.eval()
with torch.no_grad():
preds = model(batch)
for i,pred in enumerate(preds):
pred = pred.argmax(0)
pred = pred.cpu().numpy()
rescaled = (255.0 / pred.max() * (pred - pred.min())).astype(np.uint8)
im = Image.fromarray(rescaled)
im.save(f'pred_{i}.png')item_tfms = Pipeline([
PILImage.create,
RandomResizedCrop(224),
ToTensor()
], split_idx=1)First define the same item transforms that were used before. Notice that we pass in split_idx=1 to ensure they utilize the validation transform behavior.
batch_tfms = Pipeline([
IntToFloatTensor(),
Normalize.from_stats(*imagenet_stats)
])Next these are our usual batch transforms. We don’t need to pass in a split_idx here because these transforms behave the same on the training as they do on the validation sets.
batch.append(item_tfms(fname))
batch = torch.stack(batch, dim=0)
batch = batch_tfms(batch.cuda())Then we need to apply both the item and batch transforms and create a “batch” of data.
with torch.no_grad():
preds = model(batchThen we get inference on our predictions
for i,pred in enumerate(preds):
pred = pred.argmax(0)
pred = pred.cpu().numpy()
rescaled = (255.0 / pred.max() * (pred - pred.min())).astype(np.uint8)
im = Image.fromarray(rescaled)
im.save(f'pred_{i}.png')Before finally decoding and saving away our images.